Description
Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
DESCRIPTION:
This study focuses on the classification of chromosomes into 23 types, a step towards fully automatic karyotyping. This study proposes a convolutional neural network (CNN) based deep learning network to automatically classify chromosomes. The proposed method was trained and tested on a dataset containing 10304 chromosome images and was further tested on a dataset containing 4830 chromosomes. The proposed method achieved an accuracy of 92.5%, outperforming three other methods that appeared in the literature. To investigate how applicable the proposed method is to the doctors, a metric named proportion of well-classified karyotype was also designed. A result of 91.3% was achieved on this metric, indicating that the proposed classification method could be used to aid doctors in genetic disorder diagnosis. Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
EXISTING SYSTEM:
- SVM classifier
- K means clustering
DRAWBACKS:
- High Computational load
- Poor discriminatory power
- Less accuracy in classification
Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
PROPOSED SYSTEM:
- A CNN passes an image through the network layers and outputs a final class. The network can have tens or hundreds of layers, with each layer learning to detect different features. Filters are applied to each training image at different resolutions, and the output of each convolved image is used as the input to the next layer. The filters can start as very simple features, such as brightness and edges, and increase in complexity to features that uniquely define the object as the layers progress. Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
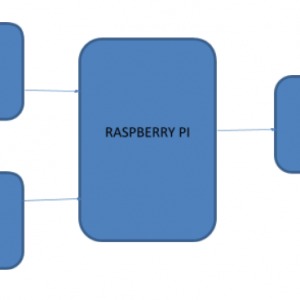
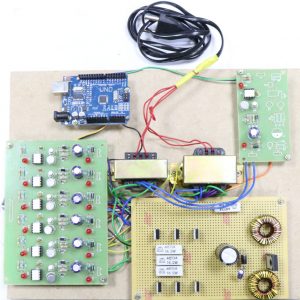
BLOCK DIAGRAM:

CNN
CNN’s, like neural networks, are made up of neurons with learnable weights and biases. Each neuron receives several inputs, takes a weighted sum over them, passes it through an activation function, and responds with an output. The whole network has a loss function and all the tips and tricks that we developed for neural networks still apply on CNN.
Multi-Band MIMO Antenna Design for 4G and 5G Mobile Terminals
SOFTWARE REQUIREMENT
PYTHON IDLE ABOVE 3.0
OPENCV
HARDWARE REQUIREMENT
SYSTEM ABOVE 4 GB RAM
CONCLUSION:
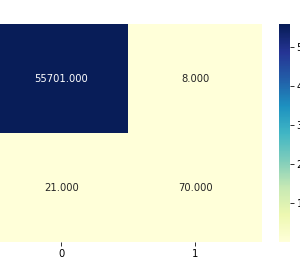
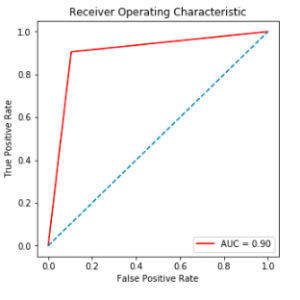
In this study, an automatic-classification method based on CNN was proposed. The model extracts chromosome images from karyotype and outputs their classes. Compared with three other methods and deep learning algorithms, our method achieved better accuracy. Our experiment also shows that our method is applicable in real-life tasks.
REFERENCES:
[1] Swati, G. Gupta, M. Yadav, M. Sharma, and L. Vig,?Siamese networks for chromosome classification,? in IEEE International Conference on Computer Vision Workshop, 2018, pp. 72? 81. [2] J. M. Cho,?Chromosome classification using backpropagation neural networks,? IEEE Engineering in Medicine and Biology Magazine, vol. 19, no. 1, pp. 28?33, 2000. [3] J. Cho, S. Y. Ryu, and S. H. Woo,?A study for the hierarchical artificial neural network model for Giemsa-stained human chromosome classification,? in International Conference of the IEEE Engineering in Medicine & Biology Society, vol. 2, 2004, pp. 4588? 4591. [4] S. Delshadpour,?Reduced size multi-layer perceptron neural network for human chromosome classification,? in International Conference of the IEEE Engineering in Medicine & Biology Society, vol. 3, 2003, pp. 2249? 2252. [5] B. C. Oskouei and J. Shanbehzadeh, ?Chromosome classification based on wavelet neural network,? in International Conference on Digital Image Computing: Techniques and Applications, 2010, pp. 605? 610. [6] S. Gagulapalalic and M. Can, ?Human chromosome classification using competitive neural network teams (can’t) and nearest neighbor,? in Ieee-Embs International Conference on Biomedical and Health Informatics, 2014, pp. 626? 629. [7] M. J. Roshtkhari and S. K. Setarehdan, ?Linear discriminant analysis of the wavelet domain features for automatic classification of human chromosomes,? in 2008 9th International Conference on Signal Processing, Oct 2008, pp. 849? 852. [8] Q. Wu, Z. Liu, T. Chen, Z. Xiong, and K. R. Castleman, ?Subspacebased prototyping and classification of chromosome images,? IEEE Transactions on Image Processing, vol. 14, no. 9, pp. 1277?1287, Sept 2005. [9] Y. Qiu, X. Lu, S. Yan, M. Tan, S. Cheng, S. Li, H. Liu, and B. Zheng, ?Applying deep learning technology to automatically identify metaphase chromosomes using scanning microscopic images: an initial investigation,? Proceedings of SPIE, vol. 9709, 2016. [10] M. Sharma, O. Saha, A. Sriraman, R. Hebbalaguppe, L. Vig, and S. Karande, ?Crowdsourcing for chromosome segmentation and deep classification,? in Computer Vision and Pattern Recognition Workshops, 2017, pp. 786? 793.































































































































































































































































































































































































































































































































































































































































































































































Customer Reviews
There are no reviews yet.